The GEO database at NCBI has a ton of data, but finding it and getting access to it can be less than straightforward. Public data is a resource that people can use to do science, or to enhance the science around a system they are currently studying in the lab, but you have to be able to find the data in order to use it.
As someone who works in many systems with many organisms, I’m often looking for either ChIP-Seq or RNA-Seq data involving a particular gene in a particular organism. There are a lot of search fields at GEO, which ones are most relevant to me? There’s a good page of examples at GEO, and some primary search fields summarized below.
Search Fields of interest:
| Thing of Interest | Examples |
|---|---|
| key words | RNA Polymerase AND stem cell |
| species | mouse[organism] |
| ChIP Seq | genome binding/occupancy profiling by high throughput sequencing[DataSet Type] |
| RNA Seq | expression profiling by high throughput sequencing[DataSet Type] |
| NGS data | high throughput sequencing[Platform Technology Type] |
| experiment variable | age[Subset Variable Type] |
Thus, if we want to form a GEO query to look for ChIP Seq experiments of RNA Polymerase in mouse, we could go to GEO and use the following search phrase:
RNA Polymerase AND stem cell AND mouse[organism] AND genome binding/occupancy profiling by high throughput sequencing [DataSet Type]
You would use an expression like that at the GEO search page. From the result, you can get the GSE and the Bioprojet number (PRJN…) or the SRA number.
The search above returned 87 records (in 2018). Each record has a GSE accession number, and a BioProject ID (PRJNAXXXXXX). These numbers can be used to in conjunction to retreive the sample id and fastq files for an experiment. You need both numbers because they each return records from different databases. The GEO DataSet database has sample names and sample ids, and the SRA database has sample ids and fastq file ids.

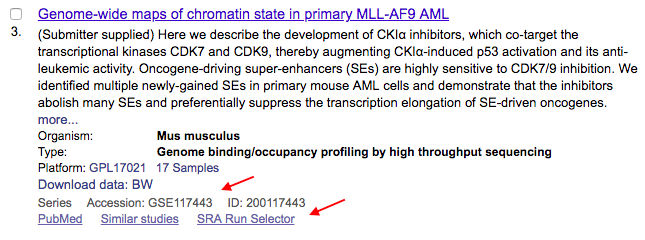
Look also at the Download data link, as it may list a file type you need, such as bigWig (BW). Otherwise, follow the steps below to download fastq files for alignment using the NCBI utility: fastq-dump.
The first step is to use the GSE number to get a file of sample information with GSM numbers. For instance, for accession: GSE87822, you can use the esearch utility to query the gds database to return a record with sample names, sample GSM numbers, and the BioProject ID:
esearch -db gds -query GSE87822 | efetch > GSE87822.txtIf you examine this file, it should have the SRA run selector in it (the PRJN number). You can extract this and use it to get the runinfo:
esearch -db sra -query PRJNA347885 | efetch --format runinfo > GSE87822_runinfo.csvFrom this file you can extract out the SRR numbers and give them to fastq-dump to download fastq files.
cut -d ',' -f1 GSE87822_runinfo.csv | grep SRR | xargs fastq-dumpYou could combine those two last steps into one, and go straight from an BioProject ID to fastq files without saving an intermediate file, but I find the intermediate information useful. The fastq files will have SRR names. The GSE87822_runinfo.csv file has those and GSM ids. You use these to link back to the sample names in the GSE87822.txt file. You have to come up with some way to join the information so you can rename your files accurately.
The fastq-dump command above will download the files to your current directory, but you can specify an output directory with --outdir, and if you have paired-end end data, you have to supply an argument to fast-dump to split the files to separate the reads with --split-files.
Lastly, if you know an SRR number, SRR4413890 for example, and just want to download it by hand, you can simply call fast-dump on the command line from the computational servers as follows:
fastq-dump SRR4413890and it will download the fastq file for that sample.
Chris Seidel, October 2018