Introduction
Fluctuation spectroscopy is a powerful method to obtain information about molecular diffusion and brightness (oligomerization). Unfortunately, interpretation of FCS results is not always straightforward given the complex nature of diffusion and photon counting noise. As a result, it is wise to learn FCS by performing simulations of molecular diffusion. I have written a plugin for simulation of FCS data in ImageJ.
The simulations work in a fairly basic way. At each time point in the simulation, molecules are moved by calculating a Gaussian random number for each dimension of free diffusion. The Gaussian random number has a standard deviation of where D is the diffusion coefficient (usually measured in um2/sec) and t is the time between simulation steps. The simulation typically takes place in a 3D box, so the positions of the molecules are initialized by calculating a uniform random number for each dimension between 0 and the length of the box. If the molecule jumps outside the dimensions of the box, it can either be reflected back into the box or reappear at the other side of the box (periodic boundary conditions).
At each point in time, it is necessary to calculate the intensity for the simulation. Here we just use the equation for the focal volume or point spread function (PSF) as follows:
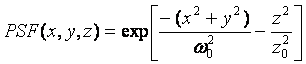
The brightness of each molecule is defined in terms of counts per second per molecule at the center of the point spread function. As a result, the above equation is simply multiplied by the brightness per sampling interval to calculate the intensity for each molecule. Typically a cutoff is applied so that molecules far from the focus are not included in the intensity. This saves considerable computational time.
In an FCS experiment, the actual intensity is not measured but rather an intensity that is blurred by shot noise. There are two options for shot noise in the simulation: poisson and analog. Poisson shot noise simulates a photon counting detector and analog shot noise simulates an analog detector detecting photocurrent. For the poisson shot noise, a poisson random number is calculated with a mean equal to the sum intensity from all the molecules in or near the PSF as described above. For analog noise, there are two contributions. Firstly, each photon results in a pulse of photocurrent. These pulses are typically exponentially distributed in amplitude. Secondly, read noise is added to the entire signal—this is typically Gaussian distributed. Therefore, the analog signal is calculated as follows: First the a poisson random number is calculated to get the number of photons detected. Then for each photon an exponential random number is calculated and summed with an average amplitude equal to the gain of the system. Finally, a Gaussian random number is added for the read noise and an offset is added as well. Therefore, the user can adjust the offset, the gain, and the read noise standard deviation. It is important to note that most detectors have non-linear gain response, so if the gain on the detector is doubled, the actual “intensity-per-photon” will increase by much more than double. The gain used in the program is the actual “intensity-per-photon” so the user must be aware of this difference.
The simulations can be set up with several different scanning modes. Point is for traditional non-scanning fcs measurements. Raster line and Raster image mimic confocal laser scanning microscopes. Line and image modes mimic a fast camera system where a snapshot of the simulation box is collected by either an entire camera or a line of the camera. Line scan image mimics a line scanning microscope.
Installing and running the plugins:
All of these plugins run within ImageJ. ImageJ can be downloaded from http://rsbweb.nih.gov/ij/download.html for free. To install the simulation plugins, copy Jay_Plugins.jar to your plugins folder (c:\Program Files\ImageJ\plugins on windows systems). Then start ImageJ and the plugins should appear under the plugins menu.
Running the plugins is relatively straightforward. Just click them from the plugins menu. The plugins will initially run with a set of parameters that works well for FCS analysis. Most other options are straightforward and I will not cover them in more detail here. The result of a simulation is a Plot Window with a trajectory (or two) displayed in it. These trajectories can be directly analyzed by the correlation and PCH plugins I have written.